当前位置:网站首页>Interpretation of the paper: iterative feature representation method to improve the prediction performance of N7 methylguanosine (m7G) sites
Interpretation of the paper: iterative feature representation method to improve the prediction performance of N7 methylguanosine (m7G) sites
2022-07-23 12:22:00 【Windy Street】
Iterative feature representation algorithm to improve the predictive performance of N7-methylguanosine sites
The article links :https://academic.oup.com/bib/article-abstract/22/4/bbaa278/5964186
DOI:https://doi.org/10.1093/bib/bbaa278
Periodical :Briefings in Bioinformatics( Area 1 )
Influencing factors :11.622
Release time :2021 year 7 month
data :http://server.malab.cn/m7G-IFL/Download.html
The server :http://server.malab.cn/m7G-IFL/
1. An overview of the article
N7- Methyl guanosine (m7G) Is an important epigenetic modification , It plays an important role in gene expression regulation . Although high-throughput experimental methods can accurately locate m7G site , But they are still cost-effective .
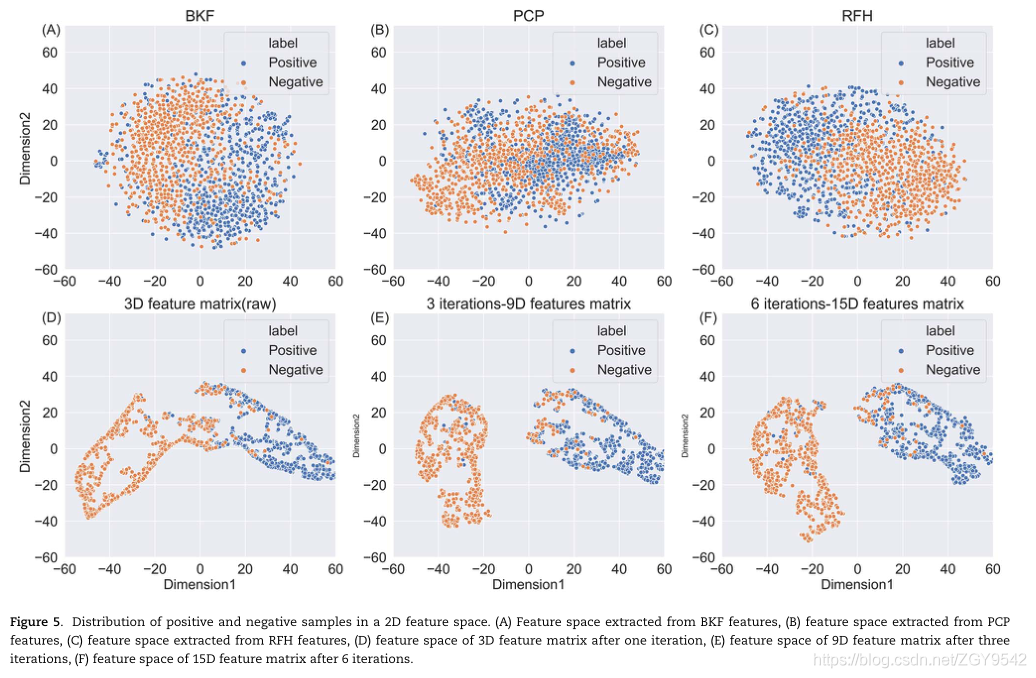
By using iterative feature representation algorithm , Developed a method based on machine learning :m7G-IFL, Used to identify m7G site . Yes m7G-IFL It is evaluated and compared with the existing prediction methods . It turns out that ,m7G-IFL Methods are identifying m7G The accuracy of loci is better than the existing prediction methods . By analyzing and comparing the characteristics used in the prediction methods , The author finds that the separation degree of positive and negative samples in the feature space is higher than that in the existing feature space . This result shows that features extract more discriminant information through iterative feature learning process , Thus, it helps to improve the prediction performance .
2. background
m7G RNA Methylation is under the action of methyltransferase , send RNA Guanine (G) Seventh place in N A modification with methyl added (N7-methylguanosine,m7G). Studies have shown that ,m7G RNA Methylation modification exists in various molecules , Include :mRNA 5’ Hat structure 、mRNA Inside 、pri-miRNA、 Transport RNA(tRNA) And ribosomes RNA(rRNA).m7G RNA Methylation modification can regulate mRNA The transcription of 、miRNA Biosynthesis and biological functions of 、tRNA stability 、18S rRNA In core processing and maturation .m7G RNA Methylation modification as a new class RNA Methylation , In the past two years, articles with high impact factors have continued , Is the m6A Another hot spot of epigenetic transcriptomics after modification .
In this study , The author puts forward m7G-IFL, This is a method for identifying m7G A new computational prediction method for loci . In this prediction method , The author introduces a method for RNA Iterative feature representation algorithm of sequence coding , The algorithm can automatically learn probability distribution information from different sequence models , And improve the ability of feature representation by supervised iteration . The author evaluates and compares our prediction method with existing prediction methods . Through feature analysis , The author finds that the proposed iterative feature algorithm can enhance the feature representation ability in the iterative process . It has also been developed. Web The server :http://server.malab.cn/m7G-IFL/.
3. data
The datasets used include 741 Positive samples and 741 A negative sample . The positive samples in this data set are real m7G site , Contains a length of 41 nt( Nucleotides ) Sequence , These sequences have been experimentally verified and derived from human HeLa and HepG2 cells . The negative sample is true or false m7G The sequence contained in the site , The same length as the positive sample . In positive and negative data sets , Sequence identity is less than 80% .
4. Method

m7G-IFL It mainly consists of three steps : Data preprocessing 、 Feature extraction and iterative feature representation .
- Divide the query sequence into the same length 41nt The subsequence .
- The generated sequence is used for feature extraction . Three feature extraction algorithms are used , Including physical and chemical properties (PCP)、 Ring function hydrogen chemical properties (RFH) And cumulative nucleotide frequency (ANF) And binary and k-mer frequency (BKF). Each sequence is converted into three types of eigenvectors , Each is based on F-score And sequential forward search (SFS) The integrated feature selection strategy is further optimized . For each subsequence , Use three optimal eigenvectors to predict its truth m7G site , And obtain three probability values , Further combined as 3D Probability eigenvector .
- The obtained probabilistic feature vector is further input into the iterative feature learning strategy , To learn the best probability eigenvector of the sequence . Last , Each sequence gets one from 0 To 1 Unequal scores . If the score is greater than 0.5, Then the sequence is predicted as m7G site ; otherwise , It is a non m7G Site .
4.1 feature extraction
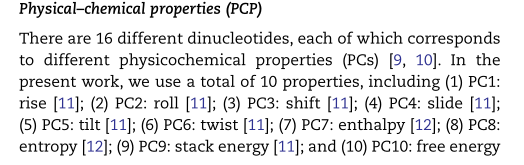
4.1.1 Physical and chemical properties (PCP)


4.1.2 Ring functional hydrogen properties (RFH) And cumulative nucleotide frequency (ANF)
Ring functional hydrogen properties (RFH):
A:(1,1,1)
C:(0,1,0)
G:(1,0,0)
U:(0,0,1)
Cumulative nucleotide frequency (ANF):
4.1.3 Binary sum k-mer frequency (BKF)
Binary system :
A:(0,0,0,1)
C:(0,1,0,0)
G:(1,0,0,0)
U:(0,0,1,0)
K-mer frequency : Use location independent k-mer Frequency to capture global sequence information , The author considers k= 2,3,4, Yes 336 (42+43+44) individual k-mer features .
4.2 Iterative features represent learning
4.2.1 Feature optimization for each feature
A two-step strategy is used to optimize the feature space derived from the three feature representation methods .
1. Calculation F-score Arrange the original features in descending order .
2. Use SFS Determine the best feature subset . stay SFS in , According to their F-score Add features in turn and train the corresponding model . When the corresponding model is 10 Fold cross validation achieved the highest accuracy , Feature subsets are considered the best .
4.2.2 Supervised probabilistic feature generation
From the above steps , The best models of three different feature groups are obtained . Each model generates a probability score for each sample in the data set , To measure the probability of prediction , Whether it is m7G Site . To avoid the probability of a single feature type , The author combines all the probabilities derived from the three models , And generate one for each sample 3D Probability eigenvector .
4.2.3 Iterative feature generation
1. Use steps 2 Generated in the 3D Vector training XGBoost Model , And then evaluate XGBoost Obtain the probability characteristics after the model . Compare the new features with the previous 3D Vector combination , Get a new one 5D Eigenvector .
2. Repeat the process again , That is, in each subsequent iteration, multi-dimensional input features are trained to obtain two-dimensional probability features , Then the input and output features are combined into the input features for the next iteration . If the performance reaches convergence , Then stop the iterative process .
5. result
5.1 Performance comparison of different classifiers


5.2 Iterative feature learning improves feature representation


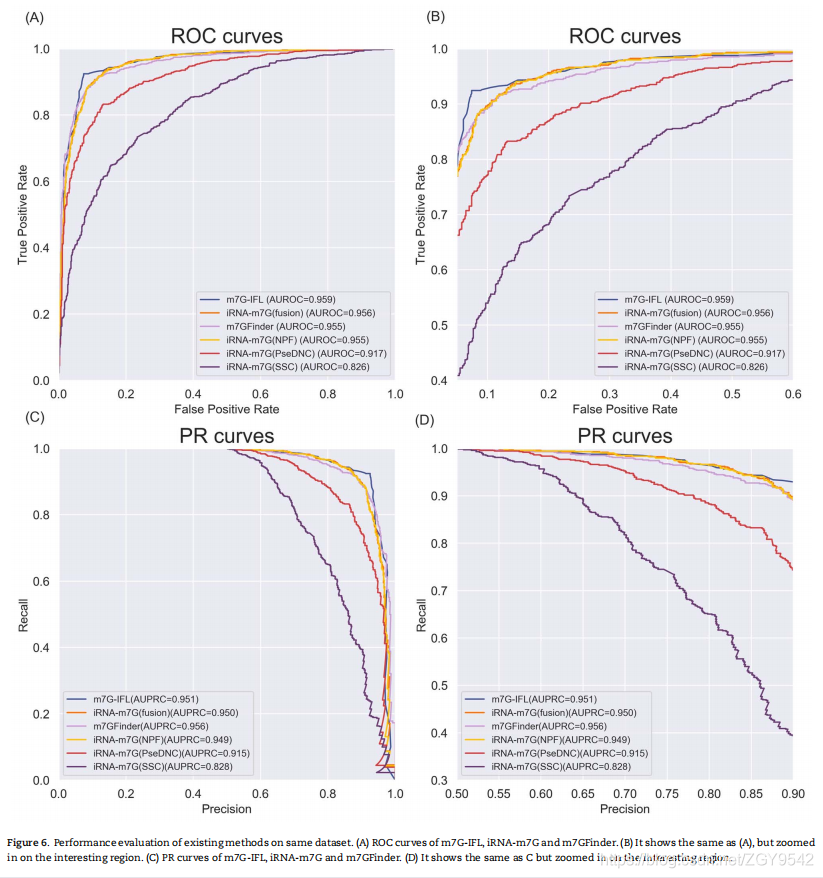
5.3 Performance comparison with existing predictors

6. summary
In this study , We have come up with a method called m7G-IFL To improve the calculation method m7G Identification of the site .
stay m7G-IFL in , The author introduces an iterative feature representation algorithm to encode RNA Sequence , The algorithm can automatically learn probability distribution information from multiple sequence models , And improve the feature representation ability in the way of supervised iteration .
Comparative studies show that , Proposed m7G-IFL Significantly better than existing ones for identifying m7G Position predictor .
The author further developed a web server , Used to implement the proposal m7G-IFL, It can provide information on the genome scale m7G High throughput prediction of loci . It can be found in http://server.malab.cn/m7GIFL/ Public access on .
边栏推荐
- 单片机学习笔记6--中断系统(基于百问网STM32F103系列教程)
- 利用pycaret:低代码,自动化机器学习框架解决回归问题
- ARM架构与编程3--按键控制LED(基于百问网ARM架构与编程教程视频)
- opencv库安装路径(别打开这个了)
- Green data center: comprehensive analysis of air-cooled GPU server and water-cooled GPU server
- Gartner调查研究:中国的数字化发展较之世界水平如何?高性能计算能否占据主导地位?
- 使用PyOD来进行异常值检测
- 2021 TOP10 development trend of information science. Deep learning? Convolutional neural network?
- High level API of propeller realizes image rain removal
- 读写文件数据
猜你喜欢

知识图谱、图数据平台、图技术如何助力零售业飞速发展

利用or-tools来求解路径规划问题(VRP)

ARM架构与编程6--重定位(基于百问网ARM架构与编程教程视频)

单片机学习笔记3--单片机结构和最小系统(基于百问网STM32F103系列教程)

论文解读:《i4mC-Deep: 利用具有化学特性的深度学习方法,对 N4-甲基胞嘧啶位点进行智能预测》

“东数西算”数据中心下算力、AI智能芯片如何发展?

Installation and use of APP automated testing tool appium

论文解读:《Deep-4mcw2v: 基于序列的预测器用于识别大肠桿菌中的 N4- 甲基胞嘧啶(4mC)位点》

《数据中心白皮书 2022》“东数西算”下数据中心高性能计算的六大趋势八大技术

Using Google or tools to solve logical problems: Zebra problem
随机推荐
How to establish data analysis thinking
Interpretation of yolov3 key code
单片机学习笔记7--SysTick定时器(基于百问网STM32F103系列教程)
硬件知識1--原理圖和接口類型(基於百問網硬件操作大全視頻教程)
The online seminar on how to help data scientists improve data insight was held on June 8
论文解读:《基于注意力的多标签神经网络用于12种广泛存在的RNA修饰的综合预测和解释》
利用or-tools来求解路径规划问题(TSP)
论文解读:《开发一种基于多层深度学习的预测模型来鉴定DNA N4-甲基胞嘧啶修饰》
How to build a liquid cooling data center is supported by blue ocean brain liquid cooling technology
数据分析的重要性
高分子物理名词解释
数据分析(一)
The data set needed to generate yolov3 from the existing voc207 data set, and the places that need to be modified to officially start the debugging program
线性规划之Google OR-Tools 简介与实战
ARM架构与编程3--按键控制LED(基于百问网ARM架构与编程教程视频)
Opencv library installation path (don't open this)
NLP自然语言处理-机器学习和自然语言处理介绍(二)
实用卷积相关trick
利用or-tools来求解带容量限制的路径规划问题(CVRP)
保存实质审查请求书出现Schema校验失败的解决方法