当前位置:网站首页>Admixture usage document Cookbook
Admixture usage document Cookbook
2022-06-27 14:57:00 【Analysis of breeding data】
The software is introduced
Genome selection , Sometimes a lot of families are measured , If you want to see the classification of these families , It can be grouped by software . Commonly used software is STRUCTURE, however STREUTURE It runs very slowly ,admixture With its computing speed , Has become the mainstream analysis software . So let's talk about that admixture How to use .
Official website
Admixture
http://software.genetics.ucla.edu/admixture/download.html

Software installation
Use conda Install the software .
conda install admixture
- 1.
After installation , type admixture, Display the following information , Description installation successful
(base) [[email protected] test]$ admixture
**** ADMIXTURE Version 1.3.0 ****
**** Copyright 2008-2015 ****
**** David Alexander, Suyash Shringarpure, ****
**** John Novembre, Ken Lange ****
**** ****
**** Please cite our paper! ****
**** Information at www.genetics.ucla.edu/software/admixture ****
Usage: admixture <input file> <K>
See --help or manual for more advanced usage.
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
- 7.
- 8.
- 9.
- 10.
- 11.
Catalog

1. Fast start
1.1 Download sample data
Be careful , The sample data on the official website can no longer be downloaded , Want to test data , You can pay attention to the official account. :“ Analysis of breeding data ”, reply “admixture”, Get test data .--------2020-5-23 to update
wget http://software.genetics.ucla.edu/admixture/hapmap3-files.tar.gz
- 1.
Once the download is complete , decompression :
tar zxvf hapmap3-files.tar.gz
- 1.
Look at the extracted file :
(base) [[email protected] admixture]$ ls
hapmap3.bed hapmap3.bim hapmap3.fam hapmap3-files.tar.gz hapmap3.map
- 1.
- 2.
Or on the official website , Download sample data : hapmap3-files.tar.gz

1.2 admixture Supported format
- plink Of bed Documents or ped file
- EIGENSTRAT The software
.geno Format
Be careful : - If your data format is plink Of bed file , such as
a.bed, Then you should include a.bim, a.fam - If your data format is plink Of ped file , such as
b.ped, Then you should include b.map
1.3 Select the appropriate number of clusters k value
Here you have to have one k value , If you don't know how many groups your group can be divided into , You can do a test , For instance from 1~7 Separate groups , Then look at their cv What's the value , Use that k value .
1.4 function k=3 Of admixture
Be careful , The name here is hapmap3.bed, instead of hapmap3( Unlike plink That doesn't add a suffix ), And there is no --file Parameters , Direct addition plink Of bed file
admixture hapmap3.bed 3
- 1.
Calculation results :
(base) [[email protected] admixture]$ admixture hapmap3.bed 3
**** ADMIXTURE Version 1.3.0 ****
**** Copyright 2008-2015 ****
**** David Alexander, Suyash Shringarpure, ****
**** John Novembre, Ken Lange ****
**** ****
**** Please cite our paper! ****
**** Information at www.genetics.ucla.edu/software/admixture ****
Random seed: 43
Point estimation method: Block relaxation algorithm
Convergence acceleration algorithm: QuasiNewton, 3 secant conditions
Point estimation will terminate when objective function delta < 0.0001
Estimation of standard errors disabled; will compute point estimates only.
Size of G: 324x13928
Performing five EM steps to prime main algorithm
1 (EM) Elapsed: 0.318 Loglikelihood: -4.38757e+06 (delta): 2.87325e+06
2 (EM) Elapsed: 0.292 Loglikelihood: -4.25681e+06 (delta): 130762
3 (EM) Elapsed: 0.29 Loglikelihood: -4.21622e+06 (delta): 40582.9
4 (EM) Elapsed: 0.29 Loglikelihood: -4.19347e+06 (delta): 22748.2
5 (EM) Elapsed: 0.29 Loglikelihood: -4.17881e+06 (delta): 14663.1
Initial loglikelihood: -4.17881e+06
Starting main algorithm
1 (QN/Block) Elapsed: 0.741 Loglikelihood: -3.94775e+06 (delta): 231058
2 (QN/Block) Elapsed: 0.74 Loglikelihood: -3.8802e+06 (delta): 67554.6
3 (QN/Block) Elapsed: 0.852 Loglikelihood: -3.83232e+06 (delta): 47883.8
4 (QN/Block) Elapsed: 1.01 Loglikelihood: -3.81118e+06 (delta): 21138.2
5 (QN/Block) Elapsed: 0.903 Loglikelihood: -3.80682e+06 (delta): 4354.36
6 (QN/Block) Elapsed: 0.85 Loglikelihood: -3.80474e+06 (delta): 2085.65
7 (QN/Block) Elapsed: 0.856 Loglikelihood: -3.80362e+06 (delta): 1112.58
8 (QN/Block) Elapsed: 0.908 Loglikelihood: -3.80276e+06 (delta): 865.01
9 (QN/Block) Elapsed: 0.852 Loglikelihood: -3.80209e+06 (delta): 666.662
10 (QN/Block) Elapsed: 1.015 Loglikelihood: -3.80151e+06 (delta): 579.49
11 (QN/Block) Elapsed: 0.908 Loglikelihood: -3.80097e+06 (delta): 548.156
12 (QN/Block) Elapsed: 0.961 Loglikelihood: -3.80049e+06 (delta): 473.565
13 (QN/Block) Elapsed: 0.855 Loglikelihood: -3.80023e+06 (delta): 258.61
14 (QN/Block) Elapsed: 0.959 Loglikelihood: -3.80005e+06 (delta): 179.949
15 (QN/Block) Elapsed: 1.011 Loglikelihood: -3.79991e+06 (delta): 146.707
16 (QN/Block) Elapsed: 0.903 Loglikelihood: -3.79989e+06 (delta): 13.1942
17 (QN/Block) Elapsed: 1.01 Loglikelihood: -3.79989e+06 (delta): 4.60747
18 (QN/Block) Elapsed: 0.85 Loglikelihood: -3.79989e+06 (delta): 1.50012
19 (QN/Block) Elapsed: 0.851 Loglikelihood: -3.79989e+06 (delta): 0.128916
20 (QN/Block) Elapsed: 0.851 Loglikelihood: -3.79989e+06 (delta): 0.00182983
21 (QN/Block) Elapsed: 0.851 Loglikelihood: -3.79989e+06 (delta): 4.33805e-05
Summary:
Converged in 21 iterations (21.788 sec)
Loglikelihood: -3799887.171935
Fst divergences between estimated populations:
Pop0 Pop1
Pop0
Pop1 0.163
Pop2 0.073 0.156
Writing output files.
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
- 7.
- 8.
- 9.
- 10.
- 11.
- 12.
- 13.
- 14.
- 15.
- 16.
- 17.
- 18.
- 19.
- 20.
- 21.
- 22.
- 23.
- 24.
- 25.
- 26.
- 27.
- 28.
- 29.
- 30.
- 31.
- 32.
- 33.
- 34.
- 35.
- 36.
- 37.
- 38.
- 39.
- 40.
- 41.
- 42.
- 43.
- 44.
- 45.
- 46.
- 47.
- 48.
- 49.
- 50.
- 51.
- 52.
- 53.
Two files will be generated :P,Q
hapmap3.3.P hapmap3.3.Q
- 1.
1.5 operation admixture when , Add error information
Add a parameter to the command summary :-B, The speed will slow down .
admixture -B hapmap3.bed 3
- 1.
Three files will be generated :P,Q,Se
1.6 If your SNP Large amount of data , Run very slowly
In choosing the best k When the value of , Can be SNP Divided into subsets , such as 50k snp It is divided into 50 A subset of , Each subset 1k SNP, Then select the best according to the subset K value , Then according to the best K It's worth running all the SNP
1.7 Multithreading
If you have multiple threads (processors), You can add parameters -jn, n Is the number of threads , Like you want to use 4 Thread run :
admixture hapmap3.bed 3 -j 4
- 1.
2. reference information
2.1 How to choose the right one K value
Multiple programs can be run at the same time , Each program is different k value , such as , to want to k It's worth choosing 1,2,3,4,5, Can be written as :
for K in 1 2 3 4 5; do admixture --cv hapmap3.bed $K | tee log${K}.out; done
- 1.
After running like this , Will generate several out file ,
hapmap3.1.P hapmap3.1.Q hapmap3.2.P hapmap3.2.Q hapmap3.3.P hapmap3.3.Q hapmap3.4.P hapmap3.4.Q hapmap3.5.P hapmap3.5.Q log1.out log2.out log3.out log4.out log5.out
- 1.
Use grep see *out Of documents cv error( The error of cross validation ) value :
grep -h CV *.out
- 1.
(base) [[email protected] admixture]$ grep -h CV *out
CV error (K=1): 0.55248
CV error (K=2): 0.48190
CV error (K=3): 0.47835
CV error (K=4): 0.48236
CV error (K=5): 0.49001
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
It can be seen that , K=3 when , CV error Minimum
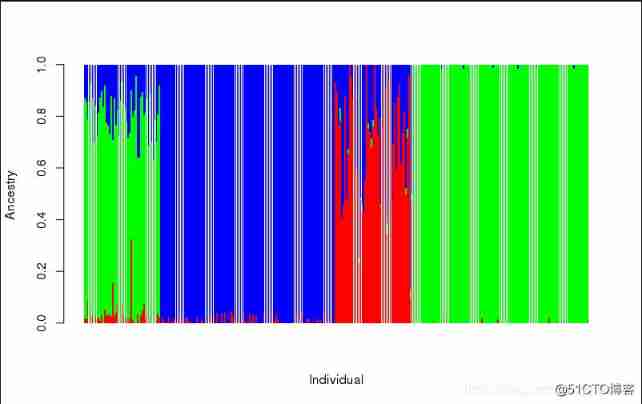
2.2 How to draw Q The chart
Use R Language
ta1 = read.table("hapmap3.3.Q")
head(ta1)
barplot(t(as.matrix(ta1)),col = rainbow(3),
xlab = "Individual",
ylab = "Ancestry",
border = NA)
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.

2.3 I need to be based on LD Get rid of some SNP Well ?
admixture Don't consider LD Information about , If you want to do this , have access to plink
such as , Here, according to plink Of bed Document carried out LD Screening
plink --bfile hapmap3 --indep-pairwise 50 10 0.1
- 1.
The filter parameter here means :
- 50, The sliding window is 50
- 10, The size of each slide is 10
- 0.1 Express R Square less than 0.1
And then it turns into bed file :
plink --bfile hapmap3 --extract plink.prune.in --make-bed --out prunedData
- 1.
The output filtered file is :
prunedData.bed prunedData.bim prunedData.fam
- 1.
Use filtered files , Run again admixture:
for K in 1 2 3 4 5 ; do admixture --cv prunedData.bed $K | tee log${K}.out;done
- 1.
(base) [[email protected] ld-test]$ grep -h CV *out
CV error (K=1): 0.52305
CV error (K=2): 0.48847
CV error (K=3): 0.48509
CV error (K=4): 0.49404
CV error (K=5): 0.49828
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
It can be seen that K=3, cv error Minimum , So choose k=3
Make a picture :
ta1 = read.table("prunedData.3.Q")
head(ta1)
barplot(t(as.matrix(ta1)),col = rainbow(3),
xlab = "Individual",
ylab = "Ancestry",
border = NA)
- 1.
- 2.
- 3.
- 4.
- 5.
- 6.
3. Other
See... For others Official pdf file
If you're interested in data analysis , For software operations , For data organization , Understanding the results , Any questions , Please feel free to contact me. .

边栏推荐
- R language objects are stored in JSON
- Naacl 2022 | TAMT: search the transportable Bert subnet through downstream task independent mask training
- Practice of constructing ten billion relationship knowledge map based on Nebula graph
- [OS command injection] common OS command execution functions and OS command injection utilization examples and range experiments - based on DVWA range
- Notes learning summary
- AbortController的使用
- CCID Consulting released the database Market Research Report on key application fields during the "14th five year plan" (attached with download)
- Design and implementation of reading app based on Web Platform
- 请求一下子太多了,数据库危
- 简析国内外电商的区别
猜你喜欢

阅读别人的代码,是一种怎样的体验

CAS之比较并交换

Getting to know cloud native security for the first time: the best guarantee in the cloud Era

How QT sets some areas to be transparent in the background image

Reflection learning summary

请求一下子太多了,数据库危

American chips are hit hard again, and another chip enterprise after Intel will be overtaken by Chinese chips
![[business security 03] password retrieval business security and interface parameter account modification examples (based on the metinfov4.0 platform)](/img/29/73c381f14a09ecaf36a98d67d76720.png)
[business security 03] password retrieval business security and interface parameter account modification examples (based on the metinfov4.0 platform)

Computer screen splitting method
Top ten Devops best practices worthy of attention in 2022
随机推荐
In the past, domestic mobile phones were arrogant in pricing and threatened that consumers would like to buy or not, but now they have plummeted by 2000 for sale
优雅的自定义 ThreadPoolExecutor 线程池
Interpretation of new version features of PostgreSQL 15 (including live Q & A and PPT data summary)
AQS抽象队列同步器
Interview question: rendering 100000 data solutions
LVI: feature extraction and sorting of lidar subsystem
Abnormal analysis of pcf8591 voltage measurement data
How is the London Silver point difference calculated
Web chat room system based on SSM
【微服务|Sentinel】热点规则|授权规则|集群流控|机器列表
做一篇人人能搞懂的ThreadLocal(源码)
Professor huangxutao, a great master in CV field, was born at the age of 86. UIUC specially set up a doctoral scholarship to encourage cutting-edge students
enable_ if
Bidding announcement: Oracle all-in-one machine software and hardware maintenance project of Shanghai R & D Public Service Platform Management Center
ERROR L104: MULTIPLE PUBLIC DEFINITIONS
Privacy computing fat offline prediction
Teach you how to realize pynq-z2 bar code recognition
Use GCC to generate an abstract syntax tree "ast" and dump it to Dot file and visualization
Library management system
NLP - monocleaner