当前位置:网站首页>J. Med. Chem. | Release: a new drug design model for deep learning based on target structure
J. Med. Chem. | Release: a new drug design model for deep learning based on target structure
2022-06-23 09:39:00 【Zhiyuan community】
This paper introduces an article from the research group of Professor Hou tingjun of Zhejiang University 、 Professor Cao Dongsheng's research group of Central South University 、 A paper jointly published by the research group of Professor lihonglin of East China University of science and technology . This paper presents a method that can take protein into account in the process of molecular formation - Deep learning generation model of ligand interaction RELATION, This model is applicable to the new drug design based on target structure .RELATION The model uses both a million - magnitude molecular library and proteins - The ligand sets data to train the variational self encoder , After introducing two-way transfer learning , The sampling of the hidden layer can take into account both the novelty of the generated molecular skeleton fragment and the affinity to the target protein .RELATION The model also provides pharmacophore constraint generation and Bayesian optimization (BO) Sampling module , It can be customized for users to generate molecules with higher matching degree of pharmacophores and better docking scoring performance for targets .

Thesis link :
https://pubs.acs.org/doi/10.1021/acs.jmedchem.2c00732

RELATION Model framework of method
边栏推荐
- 高性能算力中心 — NVMe/NVMe-oF — NVMe-oF Overview
- Sequential representation and implementation of sequencelist -- linear structure
- Set the CPU to have 16 address lines and 8 data lines, and use mreq as the access control line number Connection between memory and CPU
- Go 字符串比较
- [SUCTF 2019]CheckIn
- [GXYCTF2019]BabySQli
- Redis learning notes - geographic information location (GEO)
- 分布式锁的三种实现方式
- Three methods to find the limit of univariate function -- lobida's rule and Taylor's formula
- AI: the Elephant in Room
猜你喜欢
![[MRCTF2020]Ez_bypass](/img/cd/bd6fe5dfc3f1942a9959a9dab9e7e0.png)
[MRCTF2020]Ez_bypass

必须知道的RPC内核细节(值得收藏)!!!
Redis learning notes - AOF of persistence mechanism
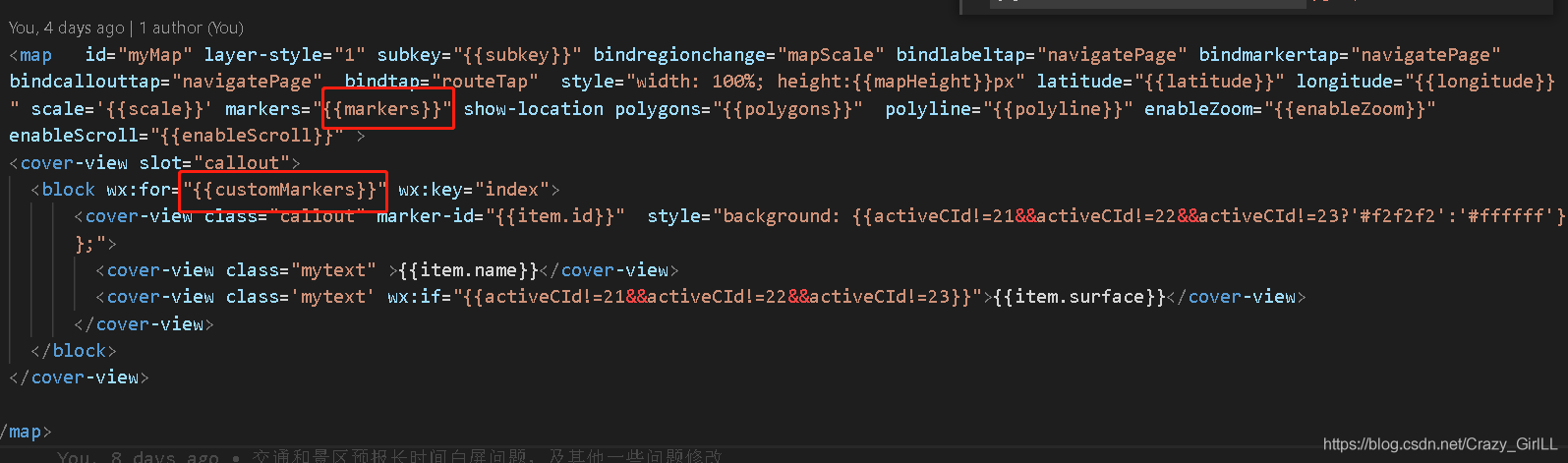
微信小程序:点击按钮频繁切换,重叠自定义markers,但是值不改变

Implementation of s5p4418 bare metal programming (replace 2ndboot)

web--信息泄漏
[nanopi2 trial experience] the first step of bare metal

swagger UI :%E2%80%8B
![[SUCTF 2019]CheckIn](/img/0e/75bb14e7a3e55ddc5126581a663bfb.png)
[SUCTF 2019]CheckIn

Cesium loading orthophoto scheme
随机推荐
UCOSII (learning notes)
Typora set up image upload service
What is a closure function
Zone d'entrée du formulaire ionic5 et boutons radio
Redis学习笔记—慢查询分析
RGB and CMYK color modes
Sun Tower Technology recruits PostgreSQL Database Engineer
ionic5錶單輸入框和單選按鈕
12个球,有一个与其他不一样,提供一个天平,三次找出来
Learn SCI thesis drawing skills (E)
学习SCI论文绘制技巧(E)
Leetcode topic analysis contains duplicate III
快速排序的简单理解
Redis learning notes master-slave copy
Pizza ordering design - simple factory model
Wechat applet: click the button to switch frequently, overlap the custom markers, but the value does not change
JSP getting started summary
Cesium加载正射影像方案
【CTF】bjdctf_ 2020_ babyrop
Leetcode topic analysis contains duplicate II

![[nanopi2 trial experience] the first step of bare metal](/img/72/fa0fca20c82853e6dbbc1f13c78100)