当前位置:网站首页>Interpretation of the paper: using attention mechanism to improve the identification of N6 methyladenine sites in DNA
Interpretation of the paper: using attention mechanism to improve the identification of N6 methyladenine sites in DNA
2022-07-23 12:08:00 【Windy Street】
Leveraging the attention mechanism to improve the identification of DNA N6-methyladenine sites
- 1. An overview of the article
- 2. background
- 3. data
- 4. Method
- 5. result
- 5.1 Quickly identify key locations
- 5.2 The similarities and differences of different levels of attention
- 5.3 AL6mA Reveals 6mA Predicted key nucleotides
- 5.4 The influence of attention level on model performance
- 5.5 Proposed model 5 Double cross validation performance
- 5.6 Performance comparison with existing methods
- 6. Key points
Article address :https://academic.oup.com/bib/article-abstract/22/6/bbab351/6359005?redirectedFrom=fulltext#supplementary-data
DOI:https://doi.org/10.1093/bib/bbab351
Periodical :Briefings in Bioinformatics( Area 1 )
Influencing factors :11.622
Release time :2021 year 8 month 28 Japan
Web The server :http://csbio.njust.edu.cn/bioinf/al6ma/
data :http://202.119.84.36:3079/al6ma/Data.html
1. An overview of the article
DNA N6- Methyladenine is an important DNA Type of embellishment , It plays an important role in many biological processes . Even though DNA Of 6mA Recent progress has been made in site prediction methods , But there are still some challenges to be solved . for example , Although manually extracted features can be explained , But it contains redundant information , This information may affect model training , And it has a negative impact on the training model . Besides , Although the model based on deep learning can automatically extract and classify features , But they lack the explicability of the key features learned by these models . therefore , A lot of research work has focused on achieving the balance between interpretability and intuition of deep learning neural networks . In this study, two-way short-term memory technology and self attention mechanism were used from DNA Extract key position information from the sequence , Two new models based on deep learning are established , For improvement N6- Methyladenine (N6-methyladenine,6mA) Prediction of loci . The performance of both methods is based on the model organism Arabidopsis (Arabidopsis thaliana) And Drosophila melanogaster (Drosophila melanogaster) Benchmarking , And make an assessment . On two benchmark datasets ,LA6mA Of AUROC Respectively 0.962 and 0.966, and AL6mA Of AUROC The areas under the curve are 0.945 and 0.941. Besides , The attention matrix is deeply analyzed , And... Hidden in the sequence are explained 6mA Important information about site prediction .
2. background
Epigenetics is considered to be an important part of genetics , It can be expressed at different levels , Including protein post-translational modification 、 RNA interfere 、DNA Embellishment, etc . In short , Epigenetics causes heritable changes in gene expression or cell phenotypes through certain mechanisms , Without changing the sequence . As a new epigenetic regulation ,DNA Methylation is found in different species , And found to be closely related to countless biological processes , Such as cell differentiation 、 Neurodevelopment and cancer suppression . Different types of methylation can occur in different positions of modification . for example ,4- Methylcytosine (4mC) It appears in the th... Of the pyrimidine ring of cytosine 4 Location ,5- Methylcytosine (5mC) It appears in the... Of pyrimidine ring 5 Location , and 6- Methyladenine (6mA) It appears in the No. of adenine ring 6 Location . In all of these mentioned above ,4mC and 5mC It has been widely studied due to their wide distribution .
For a long time , People always think that 6mA It only exists in bacteria , In eukaryotes 6mA The distribution and function of , Because it was not found in the early research . In recent years , Benefit from the progress and application of high-throughput sequencing technology , Also detected in eukaryotes 6mA.6mA Sites can be detected by a series of wet laboratory methods , These methods include but are not limited to methylation DNA Immunoprecipitation sequencing 、 Capillary electrophoresis 、 Laser induced fluorescence and pacbio Single molecule real-time sequencing . The experimental results provide a wealth of information , At the same time, there are also high costs 、 Obvious disadvantages of low efficiency .6mA Loci are unevenly distributed in the genome , At present 6mA The understanding of modification function is still very limited . therefore , Prediction by single nucleotide decomposition 6mA site , And explore the key information around the target methylation site , It is of great significance for the epigenetic regulation of gene expression and its association with human diseases .
At present, several eukaryotes have been developed 6mA Calculation method of site prediction . Early methods focused on the representation and extraction of manual features , And use the traditional machine learning algorithm to predict . for example ,chen Et al. Proposed the first one based on machine learning (ML) Methods i6mA-Pred be used for 6mA Site recognition , It uses nucleotide chemistry and nucleotide frequency as input characteristics , Combined with support vector machine (SVM) Train and predict .i6mA-DNCP Use dinucleotide composition and dinucleotide based DNA Attribute as input , combination bagging The classifier makes predictions .iDNA6mA-Rice Single nucleotide binary coding is used for sequence representation , And use random forest to classify .SDM6A Five different codes are used to identify the best feature set , Input to support vector machine and extremely random tree classifier .6mA-Finder Use recursive feature elimination strategy from 7 Sequence derived features and 3 Select the best feature group from the features based on physical and chemical properties . People often use manually extracted features , For example, the cumulative frequency of nucleotides (ANF)、k-mer frequency 、 Pseudo electron ion interaction (EIIP)、 Position specific trinucleotide propensity and pseudonucleotide composition to represent the sequence . Besides , Some methods also use statistical models . for example ,MM-6mAPred A first-order Markov model is used to identify 6mA site . Although the combination of manual features and machine learning classifiers is widely used in the processing of genome sequences , But inevitably there will be some defects . for example , Handmade features have redundancy of information , And very subjective , Although they can be explained . Besides , The classifier often ignores the hidden information in the sequence , It makes the features extracted manually difficult to be the best choice for training classifiers . With deep learning (DL) Technological development , Some sequence based end-to-end algorithms are used 6mA Site recognition . These methods include iDNA6mA (5 Step rule )、SNNRice6mA、DeepM6A、Deep6mA and i6ma-DNC, They are all based on convolutional neural networks in sequence one-hot Model encoded as input . especially ,i6ma-DNC take DNA The sequence breaks down into dinucleotide components , And then type in CNN Model , testing N6- Methyladenine site .Deep6mA take CNN and LSTM Joint prediction 6mA site , Found in different species 6mA There is a similar pattern at the site of .
In this study , The author puts forward two methods for 6mA End to end method of loci :LA6mA and AL6mA. These two sequence based methods automatically extract sequence features , With DNA The sequence is the only input , distinguish 6mA Site and non - 6mA site , Thus avoiding the tedious and over dependence of extracting manual features . Besides , Two way long-term and short-term memory (Bi-LSTM) For from DNA Capture important short-range and long-range information in the sequence , And the self attention mechanism is used to capture the position information of the sequence . Through the experiment of benchmark data , use LSTM And self attention mechanism to test the effectiveness of the method . In addition, the attention matrix is analyzed in detail , Including the key positions of the input sequence 、 The change of attention vector when paying attention to these key positions, and the similarity and difference of two models and two model biological attention layers . The author found , The difference between positive samples and negative samples at the attention level , It is helpful to understand why these models can make correct predictions . A lot of experiments show that , Proposed LA6mA and AL6mA The competitive performance of the method is better than other existing most advanced 6mA Better prediction method .LA6mA and AL6mA The online network server of has been realized .
3. data
In this study ,DNA Of 6mA The data comes from two species : Arabidopsis (Arabidopsis thaliana) And Drosophila melanogaster (Drosophila melanogaster ). The raw data comes from PacBio Public database . By excluding sequence variance upstream of the identified modification site 10bp And downstream 5bp Between 、 The variation rate of methylation level is expected to be greater than 30% The candidate genes were further screened . After screening , Respectively obtained 19632 Arabidopsis (A. thaliana) and 10653 A black bellied fruit fly (D. melanogaster ) Of 6mA site . Select the same number of non 6mA Sites as negative samples . Each non 6mA The locus is away from any adjacent 6mA At least 200 Base .
The authors further screened these sequences , Eliminate those that contain uncertainty DNA Sequence of base sites . Last , Retain the 19616 Arabidopsis (A. thaliana) Positive samples and 10653 A black bellied fruit fly (D. melanogaster ) Positive sample . For every kind of creature , Press as 9:1 The proportion is randomly divided into training set and independent test set .
4. Method
4.1 Characteristic means
The binary thermal coding scheme is adopted for the input DNA The sequence is represented : A = [1,0,0,0] ,C = [0,1,0,0] ,G = [0,0,1,0] and T = [0,0,0,0,1]. This coding scheme makes the elements in the coding matrix correspond to the bases in the input sequence , Easy attention matrix / Vector analysis . therefore , Each length is L Of DNA The sequence is converted into a size of L×4 Of 2D matrix . The length of each sequence is 41bp, By each side 20 Nucleotides and central adenine sites .
4.2 Network structure

LA6mA The skeleton of is shown in the figure 1A Shown . Its two-way LSTM The layer is first connected to the coding matrix , Two of them are bidirectional LSTM Layer of num_units = 32. And then use it LSTM Each time step of connects the attention layer , Weight coefficient matrix T∈RL×k Parameters in k = 32. Last , In full connection (FC) After the layer , Note that the layer is fixed and connected to the output .FC The number of nodes in the layer is set to 100.
AL6mA The structure of is shown in the figure 1D Shown . Pair length L Encode the input sequence of , Then connect directly to the attention layer . Note that there is a two-way LSTM layer , Its parameters are as follows : num _ units =128、time _ steps = 41. Last , Take the output of the last time step as the final prediction result .
It is worth noting that , These two proposed methods are not only used to predict potential methylation sites . They can also deeply analyze the hidden information that the model pays attention to and uses , To make predictions . chart 1B Sum graph 1C Describes how the attention matrix is analyzed and explained .
5. result
5.1 Quickly identify key locations
chart 2 It shows AL6mA and LA6mA For Arabidopsis (A. thaliana) The experimental results of , Including randomly initialized attention vectors 、 The attention vector of the final model and the change of attention vector .
chart 2C Sum graph 2F Each shows AL6mA and LA6mA Random initial attention vector of method . It can be seen that ,AL6mA The initial attention weight of seems to be randomly distributed throughout the sequence ( chart 2C) , This is obviously different from others . stay LA6mA Under the circumstances , Its initial attention weight is almost evenly distributed ( chart 2F). When the input sequence is input to an initialized LA6mA Model time ,LSTM Layer extract the features of the sequence , Then pass it to the attention layer , This is the main reason why the values in the extracted initial attention vector are evenly distributed .
chart 2A Sum graph 2D Each shows AL6mA and LA6mA The final attention vector of . Regardless of the initial distribution of attention , Finally, the weight of the central area of the attention of the model is significantly greater than that of the edge area . This shows that the central region contributes more to the prediction of the final result . Besides , The author also speculates , The contribution of the right side to the prediction is greater than that of the left side , say concretely ,[-2,9] Regions contribute the most to the prediction in terms of attention weight . From a biological point of view , Mutations in this region may affect the possibility of adenine central site methylation . Besides , Methylation changes caused by this mutation may lead to abnormal biological processes .
In order to observe the change of the attention vector , Extract attention weight in the process of model optimization . It turns out that , as time goes on , The value of the central region gradually increases , And the value of the edge area gradually decreases , It highlights that the model can automatically focus on key areas in the optimization process . It's amazing , The change of attention vector in the key position only occurs in a few epochs On , And as the iteration goes on , The value of the attention vector is constantly fine tuned , Finally reach a high value . chart 2B Sum graph 2E Verify the rapid identification of key positions related to model prediction .
5.2 The similarities and differences of different levels of attention
The training of the final attention vector is good AL6mA and LA6mA To Drosophila melanogaster (D. melanogaster ) It's shown in figure 3.
All in all ,LA6mA Method is connecting to FC Put the attention layer behind the feature extraction layer . therefore ,LA6mA The attention layer of the model focuses on the extracted features , Instead of the original sequence . Up to a point , Attention may be distracted , Become more abstract . On the other hand ,AL6mA The model connects the attention layer directly to the input matrix , Pay attention to the underlying information , Promote the discovery of key location information .
5.3 AL6mA Reveals 6mA Predicted key nucleotides

In order to analyze the attention mechanism , Author use PLogo Generate a sequence flag representation for each position in the sequence group . say concretely , In Arabidopsis (A. thaliana) And Drosophila melanogaster (D. melanogaster ) Central A The sequence around the base is detected and recognized . according to p value < 0.05 The reference height is adjusted according to the statistical significance of . Pictured 4 Shown , In two datasets ,6mA He Fei 6mA Around the site DNA There are significant differences between enriched and depleted nucleotides in the sequence .
For trained models , You can extract from each input sequence a size of L× 4 Attention matrix . Different attention matrices are obtained from different sequence inputs , The values in the attention matrix reflect the specific areas that the model pays attention to when making predictions . Pictured 5A Shown , The heat map provides a visualization of the matrix , The values are highlighted by dark or light colors , Dark color indicates the larger value of attention matrix , Light colors are the opposite .

DNA Short nucleotides widely present in the sequence , Considered functional , be called DNA Sequence motif . under these circumstances , In addition to a single sequence of key regions , The author also analyzed the results based on a set of samples . say concretely , All test samples are entered into trained AL6mA Model , And extract all attention matrices accordingly . then , choice TP and TN The attention matrix of the sample calculates the mean value of the attention matrix . The average attention matrix can be further mapped to the sequence , To represent the amount of information provided by different positions in the sequence . chart 5B、 chart 5C Sum graph 6 It shows Arabidopsis (A. thaliana) And Drosophila melanogaster (D. melanogaster ) Result .
surface 2 And table 3 Arabidopsis is listed separately (A. thaliana) And Drosophila melanogaster (D. melanogaster ) Key nucleotides . We consider a statistically meaningful cardinality .
5.4 The influence of attention level on model performance
For test sets
Pay attention to the belt of the layer al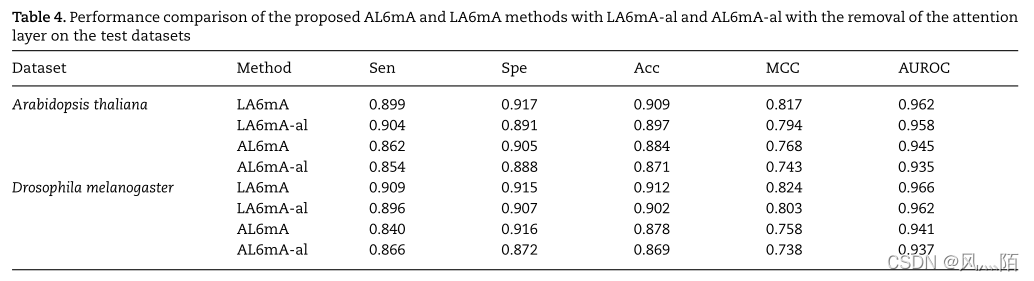
5.5 Proposed model 5 Double cross validation performance

5.6 Performance comparison with existing methods

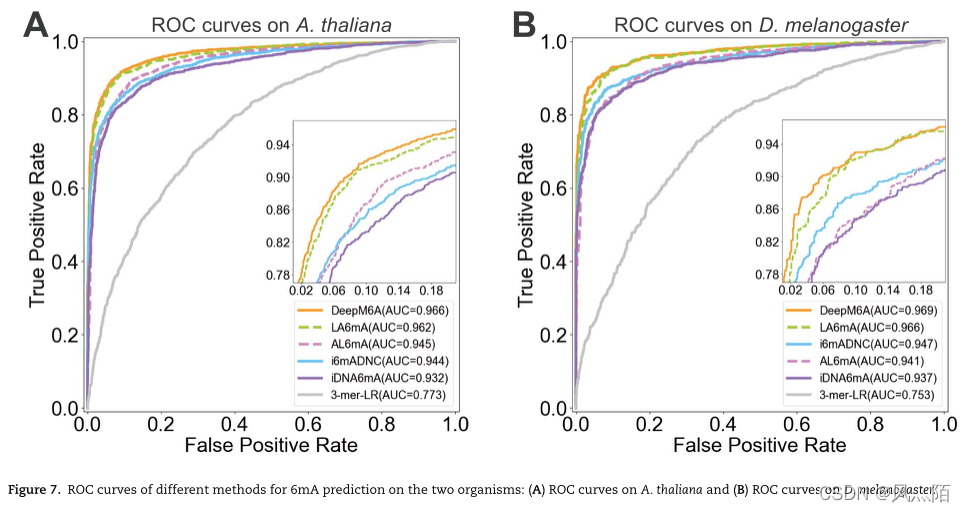
DeepM6A Is a person with 315 481 A deep convolution network of parameters , And the proposed AL6mA and LA6mA Only 138 043 individual ( Occupy DeepM6A Of 43.76%) and 159 235 individual ( Occupy DeepM6A Of 50.47%) Parameters .
6. Key points
- Accurately predict DNA Of 6mA Loci play an important role in understanding their functional roles in a variety of biological processes .
- Two new methods have been developed , Known as LA6mA and AL6mA, utilize LSTM Automatic access to DNA Information in the sequence .
- Use self attention mechanism to effectively capture DNA Location information in the sequence
- In depth analysis of two 6mA Prediction model for Arabidopsis (A. thaliana) And Drosophila melanogaster (D. melanogaster ) Changes in attention weight 、 Pay attention to the similarities and differences of layers , as well as TP and TN Pay attention to the difference of the sample layer , The key information supporting the model prediction is explained .
- Set up an online network server , This server can be used as a prediction dna 6mA Useful tools for loci .
边栏推荐
- 2021信息科学Top10发展态势。深度学习?卷积神经网络?
- The data set needed to generate yolov3 from the existing voc207 data set, and the places that need to be modified to officially start the debugging program
- APP审核期间遇到的问题总结
- UE4 solves the problem that the WebBrowser cannot play H.264
- All kinds of ice! Use paddegan of the propeller to realize makeup migration
- 1. Initial experience of MySQL
- Necessary mathematical knowledge for machine learning / deep learning
- 2. MySQL data management - DML (add, modify, delete data)
- High level API of propeller to realize face key point detection
- MySQL view
猜你喜欢

Lecturer solicitation order | Apache dolphin scheduler meetup sharing guests, looking forward to your topic and voice!

高德定位---权限弹框不出现的问题

笔记 | 百度飞浆AI达人创造营:数据获取与处理(以CV任务为主)

10、I/O 输入输出流

2、MySQL数据管理--DML(添加、修改、删除数据)

可能逃不了课了!如何使用paddleX来点人头?

Iterative display of.H5 files, h5py data operation

Data warehouse 4.0 notes - user behavior data collection I
Pytorch个人记录(请勿打开)

How to cast?
随机推荐
NT68661-屏参升级-RK3128-开机自己升级屏参
Development and deployment of steel defect detection using paddlex yolov3 of propeller
笔记 | 百度飞浆AI达人创造营:深度学习模型训练和关键参数调优详解
以不太严谨但是有逻辑的数学理论---剖析VIO之预积分
二叉树
串
High level API of propeller to realize face key point detection
Gaode positioning - the problem that the permission pop-up box does not appear
For loop
双端队列
pytorch与paddlepaddle对比——以DCGAN网络实现为例
使用飞桨的paddleX-yoloV3对钢材缺陷检测开发和部署
LearnOpenGL - Introduction
Nt68661 screen parameter upgrade-rk3128-start up and upgrade screen parameters yourself
知识图谱、图数据平台、图技术如何助力零售业飞速发展
Eigen多版本库安装
1.认识数据库
11、多线程
强迫症的硬盘分区
“东数西算”数据中心下算力、AI智能芯片如何发展?