当前位置:网站首页>Methods for converting one-dimensional data (sequence) into two-dimensional data (image) GAFS, MTF, recurrence plot, STFT
Methods for converting one-dimensional data (sequence) into two-dimensional data (image) GAFS, MTF, recurrence plot, STFT
2022-06-22 19:33:00 【xiaoweiwei99】
Methods for converting one-dimensional sequence data into two-dimensional image data detailed comprehensive
One 、 background
Although the deep learning method (1D CNN, RNN, LSTM etc. ) It can directly process one-dimensional data , But the current deep learning methods mainly deal with two-dimensional structure data , Especially in computer vision CV And natural language processing NLP field , Various methods emerge in endlessly . therefore , If one-dimensional sequence data can be transformed into two-dimensional data ( Images ) data , Can be directly combined with CV as well as NLP Domain approach , Isn't it very interesting !
Two 、 Methods to introduce
Grameen angle field GAFs
principle
take The zoom After 1D Sequence data from Rectangular coordinate system The switch to Polar coordinate system , Then by considering the angles between different points and / Difference to identify... At different points in time Time relevance . It depends on whether the angle is sum or difference , Yes There are two ways to achieve :GASF( Make an angle and ), GADF( Corresponding to the angle difference ).
Implementation steps
Step 1: The zoom , Scale the data range to [-1,1] perhaps [0, 1], The formula is as follows :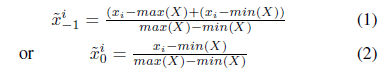
Step 2: Convert the scaled sequence data to polar coordinate system , That is, the value is regarded as the cosine value of the included angle , Time stamp is regarded as radius , The formula is as follows :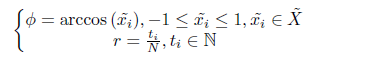
notes : If the data scaling range is [-1, 1], Then the angle range after conversion is [0, π pi π]; If the zoom range is [0, 1], Then the angle range after conversion is [0, π pi π/2].
Step 3:
You can see , Final GASF and GADF The calculation of is transformed into a rectangular coordinate system “ similar ” Operation of inner product .
The efficiency problem : For length is n The sequence data of , Converted GAFs Size is [n, n] Matrix , May adopt PAA( Piecewise aggregation approximation ) First reduce the sequence length , Then in the conversion . So-called PAA Namely : Segment the sequence , Then, the subsequence in each segment is compressed into a numerical value by averaging , Easy !
Invoke the sample
Python tool kit pytl We have provided API, in addition , The author implements the code by himself , Want to see the implementation details and get more test cases , But from my link obtain .
'''
EnvironmentPython 3.6, pyts: 0.11.0, Pandas: 1.0.3
'''
from mpl_toolkits.axes_grid1 import make_axes_locatable
from pyts.datasets import load_gunpoint
from pyts.image import GramianAngularField
# call API
X, _, _, _ = load_gunpoint(return_X_y=True)
gasf = GramianAngularField(method='summation')
X_gasf = gasf.transform(X)
gadf = GramianAngularField(method='difference')
X_gadf = gadf.transform(X)
plt.figure()
plt.suptitle('gunpoint_index_' + str(0))
ax1 = plt.subplot(121)
ax1.plot(np.arange(len(rescale(X[k][:]))), rescale(X[k][:]))
plt.title('rescaled time series')
ax2 = plt.subplot(122, polar=True)
r = np.array(range(1, len(X[k]) + 1)) / 150
theta = np.arccos(np.array(rescale(X[k][:]))) * 2 * np.pi # radian -> Angle
ax2.plot(theta, r, color='r', linewidth=3)
plt.title('polar system')
plt.show()
plt.figure()
plt.suptitle('gunpoint_index_' + str(0))
ax1 = plt.subplot(121)
plt.imshow(X_gasf[k])
plt.title('GASF')
divider = make_axes_locatable(ax1)
cax = divider.append_axes("right", size="5%", pad=0.2) # Create an axes at the given *position*=right with the same height (or width) of the main axes
plt.colorbar(cax=cax)
ax2 = plt.subplot(122)
plt.imshow(X_gadf[k])
plt.title('GASF')
divider = make_axes_locatable(ax2)
cax = divider.append_axes("right", size="5%",
pad=0.2) # Create an axes at the given *position*=right with the same height (or width) of the main axes
plt.colorbar(cax=cax)
plt.show()
The results are shown in the following figure :
Scaled sequence data and representation in polar coordinate system :
Converted GASF and GADF:
Markov transition field MTF
principle
be based on 1 Order Markov chain , Because the Markov transition matrix is not sensitive to the time dependence of the sequence , Therefore, the author puts forward the so-called MTF.
Implementation steps
Step 1: First, sequence data ( The length is n) According to its value range, it is divided into Q individual bins ( Similar to quantile ), Every data point i Belong to a unique qi ( ∈ in ∈ {1,2, …, Q}).
Step 2: Construct Markov transition matrix W, The matrix size is :[Q, Q], among W[i,j] from qi The data in is qj The frequency immediately adjacent to the data in , Its calculation formula is as follows :
w i , j = ∑ x ∈ q i , y ∈ q j , x + 1 = y 1 / ∑ j = 1 Q w i , j w_{i,j}=sum_{ orall x in q_{i}, y in q_{j},x+1=y}1/sum_{j=1}^{Q}w_{i,j} wi,j=∑x∈qi,y∈qj,x+1=y1/∑j=1Qwi,j
Step 3: Construct Markov transition field M, The matrix size is :[n, n], M[i,j] The value of is W[qi, qj]
The efficiency problem : Reason and GAFs similar , In order to improve efficiency , Try to reduce M The size of the , Ideas and PAA similar , take M Gridding , Then the subgraphs in each grid are replaced by their average values .
Invoke the sample
Python tool kit pytl We have provided API,API Interface reference :https://pyts.readthedocs.io/en/latest/generated/pyts.image.MarkovTransitionField.html. in addition , The author implements the code by himself , Want to see the implementation details and get more test cases , But from my github link obtain .
'''
EnvironmentPython 3.6, pyts: 0.11.0, Pandas: 1.0.3
'''
from mpl_toolkits.axes_grid1 import make_axes_locatable
from pyts.datasets import load_gunpoint
from pyts.image import MarkovTransitionField
## call API
X, _, _, _ = load_gunpoint(return_X_y=True)
mtf = MarkovTransitionField()
fullimage = mtf.transform(X)
# downscale MTF of the time series (without paa) through mean operation
batch = int(len(X[0]) / s)
patch = []
for p in range(s):
for q in range(s):
patch.append(np.mean(fullimage[0][p * batch:(p + 1) * batch, q * batch:(q + 1) * batch]))
# reshape
patchimage = np.array(patch).reshape(s, s)
plt.figure()
plt.suptitle('gunpoint_index_' + str(k))
ax1 = plt.subplot(121)
plt.imshow(fullimage[k])
plt.title('full image')
divider = make_axes_locatable(ax1)
cax = divider.append_axes("right", size="5%", pad=0.2)
plt.colorbar(cax=cax)
ax2 = plt.subplot(122)
plt.imshow(patchimage)
plt.title('MTF with patch average')
divider = make_axes_locatable(ax2)
cax = divider.append_axes("right", size="5%", pad=0.2)
plt.colorbar(cax=cax)
plt.show()
The result is shown in the figure :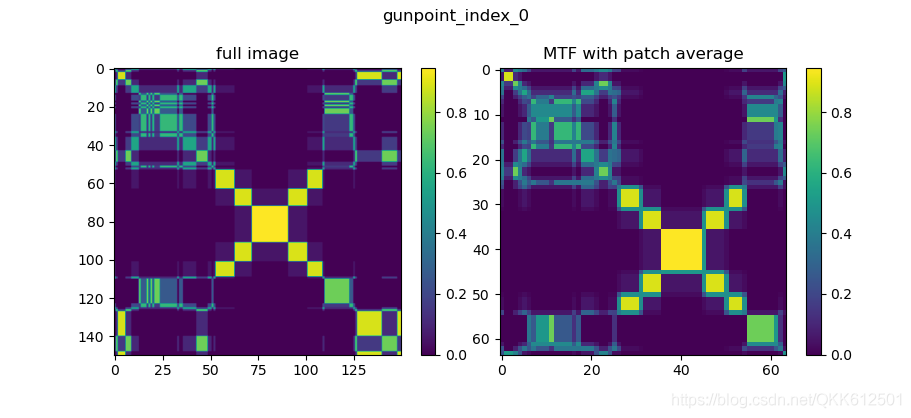
Recursive graph Recurrence Plot
Recursive graph (recurrence plot,RP) Is to analyze the periodicity of time series 、 An important method of chaos and nonstationarity , It can reveal the internal structure of time series , Give information about similarity 、 A priori knowledge of information and predictability , Recursive graph is especially suitable for short time series data , It can test the stationarity of time series 、 Internal similarity .
principle
A recursive graph is an image that represents the distance between tracks extracted from the original time series
Given time series data : ( x 1 , … , x n ) (x_1, ldots, x_n) (x1,…,xn), The extracted track is :
x i = ( x i , x i + τ , … , x i + ( m 1 ) τ ) , i ∈ { 1 , … , n ( m 1 ) τ } ec{x}_i = (x_i, x_{i + au}, ldots, x_{i + (m - 1) au}), quad orall i in {1, ldots, n - (m - 1) au } x i=(xi,xi+τ,…,xi+(m1)τ),i∈{1,…,n(m1)τ}
among : m m m Is the dimension of the trajectory , τ au τ Is delay . Recursive graph R Is the pairwise distance between tracks , The calculation is as follows :
R i , j = Θ ( ε ∥ x i x j ∥ ) , i , j ∈ { 1 , … , n ( m 1 ) τ } R_{i, j} = Theta(arepsilon - | ec{x}_i - ec{x}_j |), quad orall i,j in {1, ldots, n - (m - 1) au } Ri,j=Θ(ε∥x ix j∥),i,j∈{1,…,n(m1)τ}
among , Θ Theta Θ by Heaviside function , and ε arepsilon ε It's the threshold .
Invoke the sample
'''
EnvironmentPython 3.6, pyts: 0.11.0, Pandas: 1.0.3
'''
from mpl_toolkits.axes_grid1 import make_axes_locatable
from pyts.datasets import load_gunpoint
from pyts.image import RecurrencePlot
X, _, _, _ = load_gunpoint(return_X_y=True)
rp = RecurrencePlot(dimension=3, time_delay=3)
X_new = rp.transform(X)
rp2 = RecurrencePlot(dimension=3, time_delay=10)
X_new2 = rp2.transform(X)
plt.figure()
plt.suptitle('gunpoint_index_0')
ax1 = plt.subplot(121)
plt.imshow(X_new[0])
plt.title('Recurrence plot, dimension=3, time_delay=3')
divider = make_axes_locatable(ax1)
cax = divider.append_axes("right", size="5%", pad=0.2)
plt.colorbar(cax=cax)
ax1 = plt.subplot(122)
plt.imshow(X_new2[0])
plt.title('Recurrence plot, dimension=3, time_delay=10')
divider = make_axes_locatable(ax1)
cax = divider.append_axes("right", size="5%", pad=0.2)
plt.colorbar(cax=cax)
plt.show()
The result is shown in Fig. :
The short-time Fourier transform STFT
STFT It can be regarded as a way to quantify the time-varying frequency and phase content of non-stationary signals ..
principle
By adding window functions ( The length of the window function is fixed ), First, the time domain signal is windowed , The original time domain signal is divided into multiple segments by sliding window , And then, for each segment FFT Transformation , Thus the time spectrum of the signal is obtained ( Time domain information is preserved ).
Implementation steps
Suppose the length of the sequence is T T T, τ au τ Is the window length , s s s Is the sliding step size ,W Representation window function , be STFT It can be calculated as :
S T F T ( τ , s ) ( X ) [ m , k ] = ∑ t = 1 T X [ t ] W ( t s m ) e x p { j 2 π k / τ ( t s m ) } STFT^{( au,s)}(X)_{[m,k]}=sum_{t=1}^{T}X_{[t]} cdot W(t-sm)cdot exp{-j2pi k / au cdot (t-sm)} STFT(τ,s)(X)[m,k]=∑t=1TX[t]W(tsm)exp{j2πk/τ(tsm)}
Transformed STFT Size is :[M, K], M Represents the time dimension ,K Represents the frequency amplitude ( Plural form ), For convenience , hypothesis s = τ s= au s=τ, That is, there is no overlap between windows , be
M = T / τ M=T/ au M=T/τ,
K = τ K =lfloor au floor K=τ/2 + 1
notes : Compared with DFT, STFT To some extent, it helps us restore the time resolution , However, there is a trade-off between achievable temporal resolution and frequency , That's what's called The principle of uncertainty . say concretely , Width of window ( τ au τ) The bigger it is , The higher the frequency domain resolution , Accordingly , The lower the time domain resolution ; Width of window ( τ au τ) The smaller it is , The lower the frequency-domain resolution , Accordingly , The higher the time domain resolution .
Invoke the sample
python package scipy Provide STFT Of API, For details of the official documents, see :https://docs.scipy.org/doc/scipy/reference/generated/scipy.signal.stft.html
scipy.signal.stft(x,fs = 1.0,window =‘hann’,nperseg = 256,noverlap = None,nfft = None,detrend = False,return_oneside = True,boundary
=‘zeros’,padded = True,axis = -1 )
Parameter interpretation :
x: Time domain signal ;
fs: Sampling frequency of the signal ;
window: Window function ;
nperseg: Window function length ;
noverlap: Overlap length of adjacent windows , The default is 50%;
nfft: FFT The length of , The default is nperseg. If it is greater than nperseg Will automatically zero fill ;
return_oneside : True Returns the real part of a complex number ,None Returns the complex number .
Sample code :
"""
@author: masterqkk, [email protected]
Environment:
python: 3.6
Pandas: 1.0.3
matplotlib: 3.2.1
"""
import pickle
import numpy as np
import matplotlib.pyplot as plt
import scipy.signal as scisig
from mpl_toolkits.axes_grid1 import make_axes_locatable
from pyts.datasets import load_gunpoint
if __name__ == '__main__':
X, _, _, _ = load_gunpoint(return_X_y=True)
fs = 10e3 # sampling frequency
N = 1e5 # 10 s 1signal
amp = 2 * np.sqrt(2)
time = np.arange(N) / float(fs)
mod = 500 * np.cos(2 * np.pi * 0.25 * time)
carrier = amp * np.sin(2 * np.pi * 3e3 * time + mod)
noise_power = 0.01 * fs / 2
noise = np.random.normal(loc=0.0, scale=np.sqrt(noise_power), size=time.shape)
noise *= np.exp(-time / 5)
x = carrier + noise # signal with noise
per_seg_length = 1000 # window length
f, t, Zxx = scisig.stft(x, fs, nperseg=per_seg_length, noverlap=0, nfft=per_seg_length, padded=False)
print('Zxx.shaope: {}'.format(Zxx.shape))
plt.figure()
plt.suptitle('gunpoint_index_0')
ax1 = plt.subplot(211)
ax1.plot(x)
plt.title('signal with noise')
ax2 = plt.subplot(212)
ax2.pcolormesh(t, f, np.abs(Zxx), vmin=0, vmax=amp)
plt.title('STFT Magnitude')
ax2.set_ylabel('Frequency [Hz]')
ax2.set_xlabel('Time [sec]')
plt.show()
Running results :
obtain STFT The resulting size is :
Zxx.shaope: (501, 101), The number of frequency components is 1000 lfloor 1000 floor 1000/2 + 1 = 501, The length of the window fragment is 1e5/1000 + 1=101 ( Here should be a pad)
References
1.Imaging Time-Series to Improve Classification and Imputation
2.Encoding Time Series as Images for Visual Inspection and Classification Using Tiled Convolutional Neural Networks
3.J.-P Eckmann, S. Oliffson Kamphorst and D Ruelle, “Recurrence Plots of Dynamical Systems”. Europhysics Letters (1987)
4.Stoica, Petre, and Randolph Moses,Spectral Analysis of Signals, Prentice Hall, 2005
5.https://laszukdawid.com/tag/recurrence-plot/
summary
Finally, a link to download the time series data set is attached :http://www.cs.ucr.edu/~eamonn/time_series_data/, It contains almost all current data sets in this field .
I hope I can help you , To be continued . Welcome to exchange :[email protected]
边栏推荐
- 插槽里如何判断text为数组
- Implementing Domain Driven Design - using ABP framework - solution overview
- 小波变换db4进行四层分解及其信号重构—matlab分析及C语言实现
- 结构型模式之装饰者模式
- How much do you know about the bloom filter and cuckoo filter in redis?
- Thread pool: reading the source code of threadpoolexcutor
- Flutter series - build a flutter development environment
- 结构型模式之代理模式
- 集群、分布式、微服务概念和区别
- Message Oriented Middleware (I) MQ explanation and comparison of four MQS
猜你喜欢

Detailed explanation of session mechanism and related applications of session

5G 短消息解决方案

Pull down refresh and pull up to load more listviews

2年狂赚178亿元,中国游戏正在“收割”老外

Message Oriented Middleware (I) MQ explanation and comparison of four MQS

常用技术注解

函数的导数与微分的关系

Niuke.com: judge whether it is palindrome string

3GPP 5G R17标准冻结,RedCap作为重要特性值得关注!

Active directory user logon Report
随机推荐
实验七 触发器
结构型模式之适配器模式
2022 operation of simulated examination platform for examination question bank of welder (elementary) special operation certificate
使能伙伴,春节重大保障“不停歇”
数字赋能机械制造业,供应链协同管理系统解决方案助力企业供应链再升级
Shell programming specification and variables
Niuke network: minimum coverage substring
2022 R2 mobile pressure vessel filling test question simulation test platform operation
SSH password free login
Longest common subsequence
Aiops intelligent operation and maintenance experience sharing
jniLibs.srcDirs = [‘libs‘]有什么用?
函数的导数与微分的关系
Is flush easy to open an account? Is it safe to open a mobile account?
实验4 NoSQL和关系数据库的操作比较
jniLibs. Srcdirs = ['LIBS'] what's the use?
Flutter series - build a flutter development environment
插槽里如何判断text为数组
助力客户数字化转型,构建全新的运维体系
IPLOOK和思博伦通信建立长期合作